For anomaly likelihood I used the code mentioned here in hotgym example and the computed likelihood was always constant:
Does this ^ need to be performed outside the loop?
What data do I need to pass to this function? timestamp is current time associated to current data I assume. But what do I pass as timestep?
Unfortunately, even after updating the likelihood to be calculated correctly, the anomaly detection fails to catch anomalies:
First of all, congrats on getting this working! 
I don’t see any time patterns in this data, which is probably why it’s not finding anomalies. It looks like the time encoding is not useful in this case. You can probably remove it. It may not help, but it shouldn’t hurt.
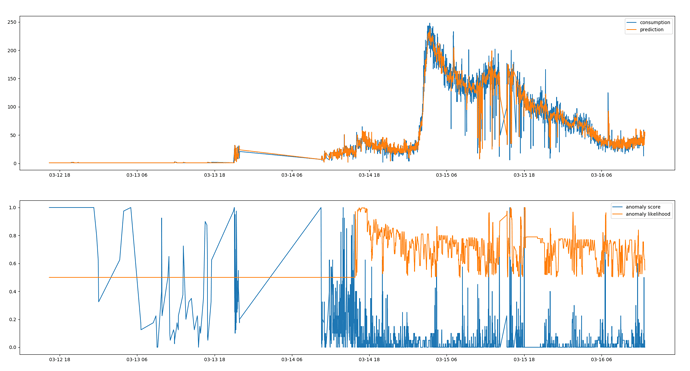
There are seasonal patterns in the data but they are only visible when I plot the entire data range:
There are multiple spikes in the data and anomaly likelihood is not pointing at any of them. Actually there are many cases where anomaly likelihood is high yet there is no obvious anomaly at all.
I guess this is game over since I don’t know what I could do to make this work. The spikes are detected correctly in HTM Studio so in theory it should work here too.
HTM Studio processes the data and chooses appropriate encoders for time as well as scalar value. We just need to find the right time encoding. Here is the code for it:
Yes as you said.
You could pass a datetime, a time step (like index val) or even nothing and it should still output likelihood values.
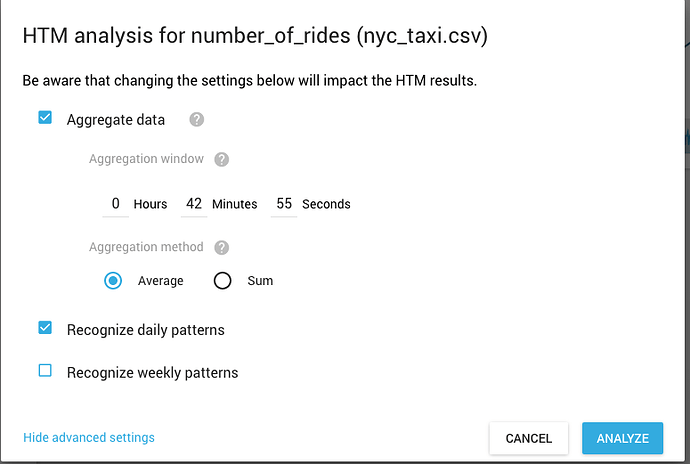
@Razvan_Flavius_Panda I just realized (thanks @lscheinkman) that you can get model aggregation settings from HTM Studio for any data you run through it. So you do not have to write the code to compute model params if you liked what you got from HTM Studio. There is an option under “show advanced settings”:
This gives you the data aggregation settings you should user for your data. You can use this in combination with getScalarMetricWithTimeOfDayAnomalyParams() to get the same model params HTM Studio used.
Also related to Using findParameters instead of swarming.
Is this the function being used by HTM Studio to find the egg window? (https://github.com/numenta/numenta-apps/blob/master/unicorn/py/unicorn_backend/param_finder.py)
Idk who wrote it but I wondered if anyone knew the basic pseudocode logic of this? Just looking for the essence of how it evaluates the different age levels, and having trouble getting it for some reason 
def _determineAggregationWindow(timeScale,
cwtVar,
thresh,
samplingInterval,
numDataPts):
"""
Determine data aggregation window
@param timeScale (numpy array) corresponding time scales for wavelet coeffs
@param cwtVar (numpy array) wavelet coefficients variance over time
@param thresh (float) cutoff threshold between 0 and 1
@param samplingInterval (timedelta64), original sampling interval in ms
@param numDataPts (float) number of data points
@return aggregationTimeScale (timedelta64) suggested sampling interval in ms
"""
assert samplingInterval.dtype == numpy.dtype("timedelta64[ms]")
assert timeScale.dtype == numpy.dtype("timedelta64[ms]")
samplingInterval = samplingInterval.astype("float64")
cumulativeCwtVar = numpy.cumsum(cwtVar)
cutoffTimeScale = timeScale[numpy.where(cumulativeCwtVar >= thresh)[0][0]]
aggregationTimeScale = cutoffTimeScale / 10.0
assert aggregationTimeScale.dtype == numpy.dtype("timedelta64[ms]")
aggregationTimeScale = aggregationTimeScale.astype('float64')
if aggregationTimeScale < samplingInterval:
aggregationTimeScale = samplingInterval
# make sure there is at least 1000 records after aggregation
if numDataPts < MIN_ROW_AFTER_AGGREGATION:
aggregationTimeScale = samplingInterval
else:
maxSamplingInterval = (float(numDataPts) / MIN_ROW_AFTER_AGGREGATION
* samplingInterval)
if aggregationTimeScale > maxSamplingInterval > samplingInterval:
aggregationTimeScale = maxSamplingInterval
aggregationTimeScale = numpy.timedelta64(int(aggregationTimeScale), "ms")
return aggregationTimeScaleWritten by @ycui, who has since left the company for work in socal. It is super complicated, I don’t understand it.
![]() Well that makes me feel better hahaha. I assume @ycui isn’t active on the forum anymore right? Otherwise do you have any intuitions for evaluating preprocessing setups for NuPIC? Of course there’s just seeing how well it does detecting known anomalies, just curious for any other ideas.
Well that makes me feel better hahaha. I assume @ycui isn’t active on the forum anymore right? Otherwise do you have any intuitions for evaluating preprocessing setups for NuPIC? Of course there’s just seeing how well it does detecting known anomalies, just curious for any other ideas.