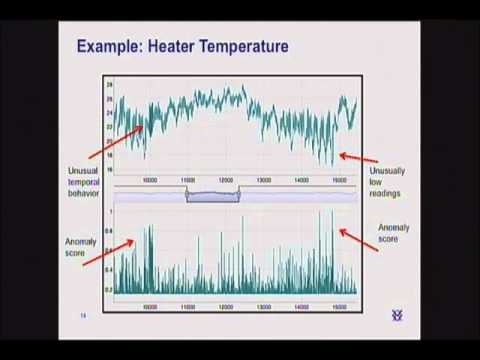
Personally, I’ve seen so many anomaly spikes (ie anomaly ratings reaching 1.0 aka 100%) that I’ve become used to seeing them quite often. You’re right, there is indeed a lot of structure to your pattern, however, I’ve understood with time that HTM cannot do inductive reasoning on patterns.
Example: say that you had a data set such as [1, 2, 8, 9, 15, 14, 1, 2, 8, 9, 15, 14, …], if further along the data your values suddenly changed, but the overall pattern remained the same such as […, 4, 5, 11, 12, 18, 17], then you would get anomaly ratings of about 1.0 at every of those values. Additionally, it would not be able to correctly predict any of those values, such as [4, 5, 11, 12, 18, ?] even though all those values followed the exact same pattern as the previous pattern, HTM would not be able to predict 17. So I don’t know what your exact EKG values are, but if there is some deviation in the actual values that is significant enough to place those values in other (maybe previously unused) buckets, then HTM will ‘ring the alarm’ and give big anomalies even though the pattern is always the same. With time however, HTM will most likely learn more of the pattern with a variety of different values and thus not give as big anomaly ratings.
However, HTM has a limit when it comes to creating what I see as “sequential memory links”, as in the previous example, HTM learned the pattern 1, 2, 8, 9, 15, 14, which means that “links” were created between the values 1 and 2, 2 and 8 and so on. But what if you had a pattern such as [1, 2, 1, 3, 1, 4, 1, 5, 1, 6, 1, 7, …] here HTM would create links between the values 1 and 2, 2 and 1, 1 and 3, 3 and 1, and so on. At some point no more new links could be created between 1 and X. Well, the main factor to influence this ability seems to be the number of cells per column (according to my tests and understanding). The more cells per column (cpc), the more cells there are to represent a value, and thus create links between that cell and cells of another column value. So the more cpc the more links can be created between values, and thus the more patterns can be learned between often used values. By default, the model_params.py sets the value of cpc to 32, and interestingly enough, after having done my tests on the mentioned example [1, 2, 1, 3, 1, 4, …, 1, X] I found X to be 14, and I had to increase the cpc value to 128 before I was able to get HTM to learn the pattern with X being 15 (haven’t tested its limits though). My point is that depending on the nature of your data, you might have to play a bit around with cpc to see if that helps HTM in learning (I know right, you must be realizing that there is quite a bit of fiddling involved here to get the optimal results).
My template uses the default encoder set by the swarm once the the model_params.py is generated, which means that is a simple ScalarEncoder. However, I usually like to manually edit the model_params.py file to RandomDistributedScalarEncoder if I get bad encoder results. [quote=“Addonis, post:7, topic:735”]
By the way, it’s funny how the HTM School video…
[/quote]
I agree  lucky coincidence or fate?[quote=“Addonis, post:7, topic:735”]
lucky coincidence or fate?[quote=“Addonis, post:7, topic:735”]
I wish there some kind of an online class about HTM Theory. Maybe in the future…
[/quote]
Time will tell  [quote=“Addonis, post:7, topic:735”]
[quote=“Addonis, post:7, topic:735”]
After running another swarm I think there is a clear difference between large and medium swarms and how much data they swarm over.
[/quote]
Not surprised, I have never run a large swarm myself, as I’m afraid it would take much too long, but I guess the reward might be worth the wait in some cases.