First off, kudos to both of you for having this conversation while I was away. Thanks @Setus for helping @Addonis out!
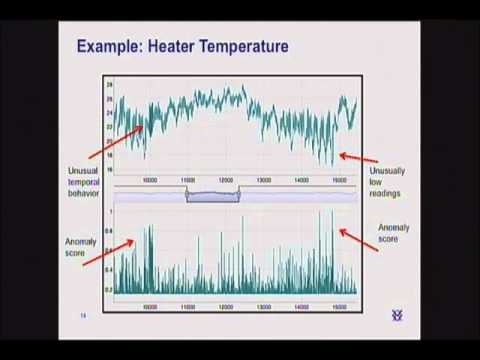
Try running the anomaly likelihood instead of the raw anomaly score. This is not actually a part of the NuPIC model, but a post-process:
As shown in the code above, you create an instance of the anomaly likelihood class and it maintains state. You pass it the value, anomaly score, and timestamp for each point and it will return a more stable anomaly indication than the raw anomaly score.
Here is a detailed explanation of the anomaly likelihood (with awful audio and video, sorry):
See Preliminary details about new theory work on sensory-motor inference.